Analytics Software
The local server-based analytics software for precision diagnostic panels provides accurate analysis results
regarding various genetic mutations based on verified open databases and in-house databases.
NGeneAnalySys®
NGeneAnalySys® is an analysis software program designed to interpret genetic variations and generate reports through the automated pipeline of data generated through the NGS equipment. For each panel, an optimized pipeline is established for variant analysis. Variants are detected based on more than 20 open public databases as well as a self-developed internal database, and the information is provided to help interpret variants.

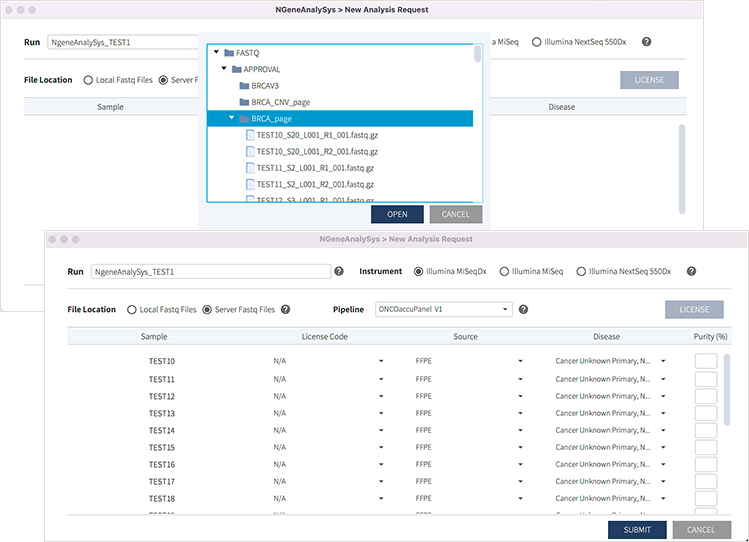
Interlinking with the NGS equipment
Data analysis is possible through linkage with the server where the data generated by sequencing equipment is stored.
When a panel is selected, the appropriate analysis pipeline is applied automatically to the analysis process.
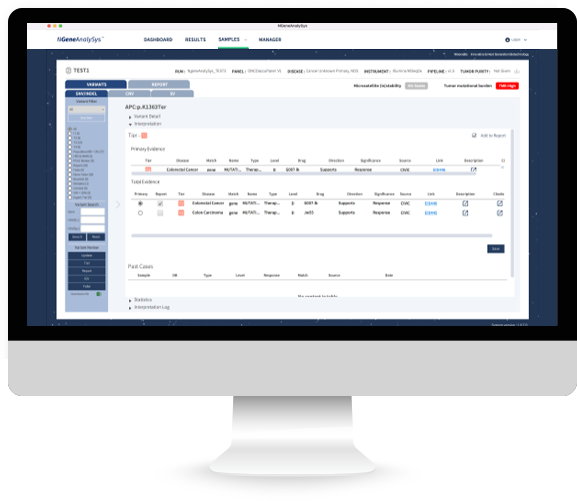
Provide mutation information and evidence according to guidelines
Tier judgment results and evidence provided according to ASCO/CAP/AMP guidelines for NGS clinical application
Tier can be modified and logs for changes are provided ※ For NGS clinical application, ACMG standards and guidelines are complied.

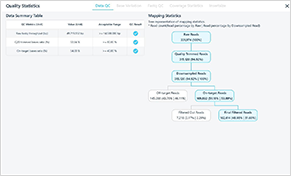
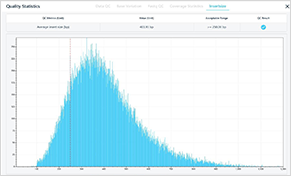
Analysis reports are provided (QC report, clinical report, etc.)
For precision medicine, various genetic databases are provided. Applying contamination prediction algorithm and in silico prediction method, intuitive and accurate variant interpretation results are made available.
WORKING PROCESS
-
- Login
-
The Client Tool is installed on the user's PC.
Login using ID and PW.
-
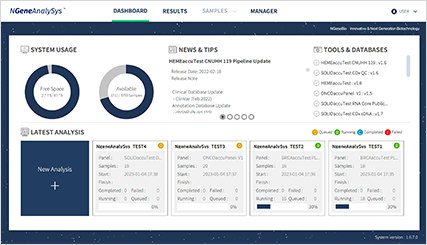
- DATA upload
-
Select data generated from the NGS equipment and upload them to the analysis server.
Once uploading is completed, analysis is initiated.
The analysis is implemented automatically by panel type selection.
The current status of the analysis is displayed.
-
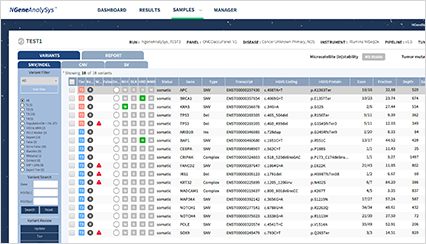
- NGS analysis
-
The list of variants detected through automatic analysis is displayed.
It is possible to check and modify classification results based on the importance of each variant.
Variant filtering based on the annotation information
-

- Report
-
Clinical Report preparation
Patient and test information check
Check detected variant type and importance
Report modification based on the review results
Final confirmation and report publication
Featured compatible panels
NGeneAnlaySys® makes available various types of related information as well as genetic variant data related to about 35 types of solid cancer including hereditary breast/ovarian cancer, hematologic malignancy, lung cancer, stomach cancer, etc.
-

BRCAaccuTest™
BRCAaccuTest™ PLUS
-

HEMEaccuTest™ DNA
HEMEaccuTest™ RNA
-

SOLIDaccuTest™ DNA
SOLIDaccuTest™ DNA HRD
SOLIDaccuTest™ RNA
-

ONCOaccuPanel™
EasyHLAanalyzer™
EasyHLAanalyzer™ is an analysis software that performs HLA genotyping by applying a bioinformatics algorithm to NGS data generated based on IMGT/HLA DB.
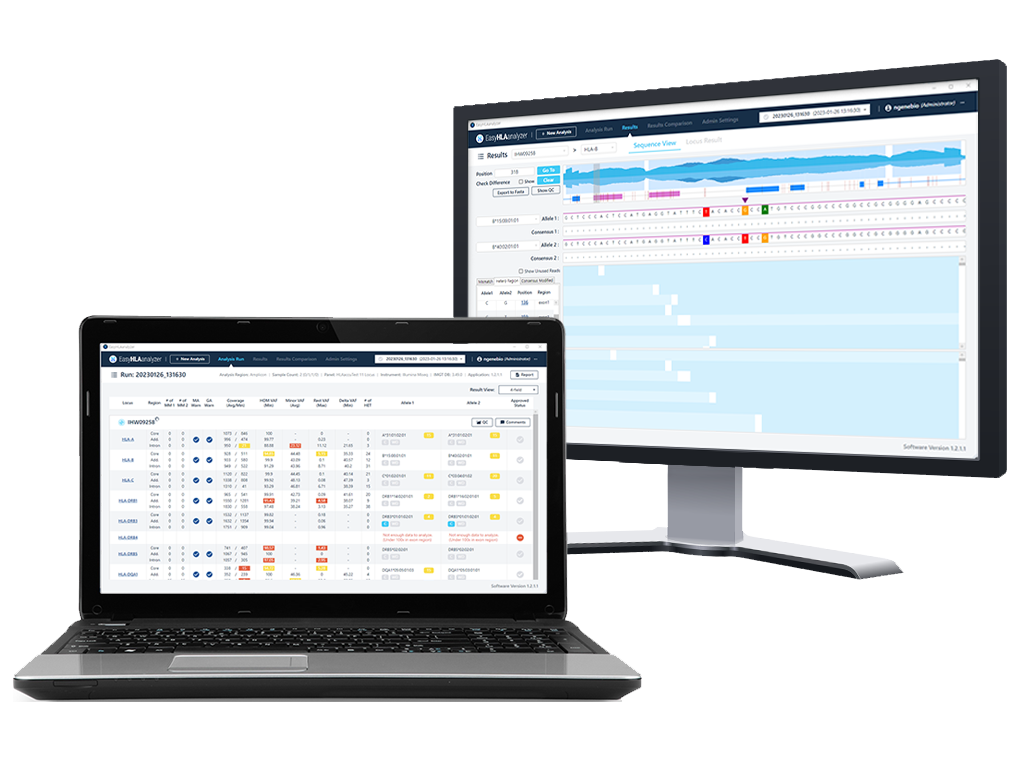
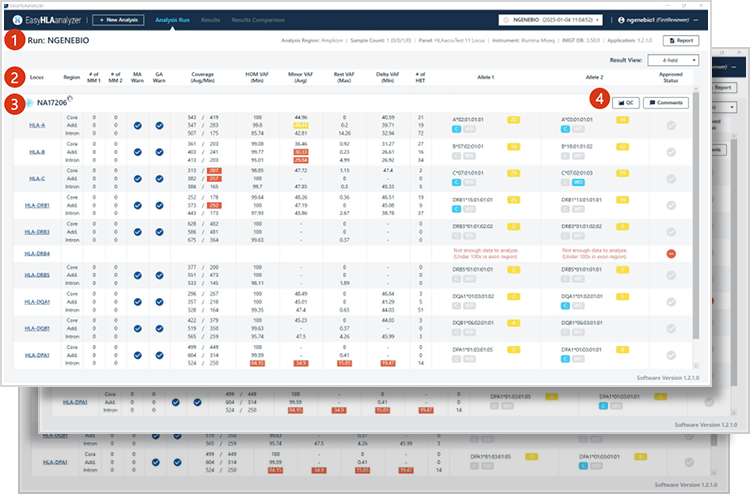
Provide database-based results for each HLA type
-
1
Run name information
Analysis Region info.
Sample number
Panel info.
NGS equipment info.
IMGT DB version
SW version
-
2
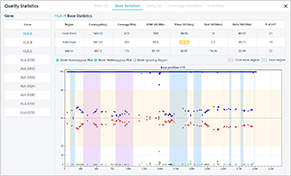
Results of sample analysis
Analyzed genes
Mismatches and warnings by Core/Add./Intron areas, Coverage, VAF, HET position info.
Allele1/Allele2 genotype
Review on analysis results
-
3
Sample Name
-
4
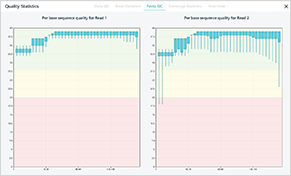
Sample QC result
Analysis result check via the genome browser
-
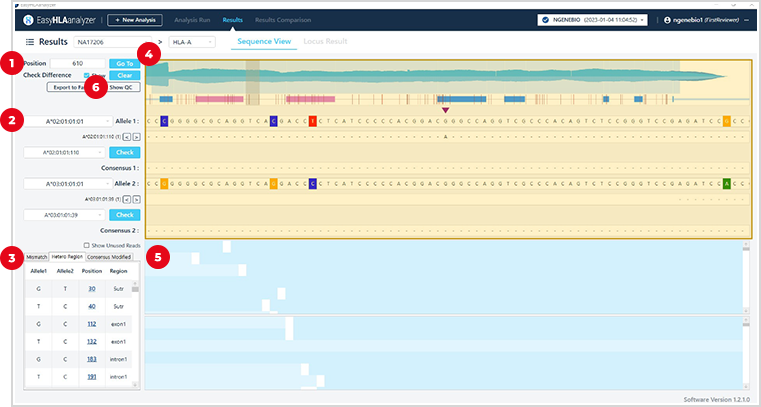
1
Gene Position
-
2
Genotype result
-
3
Mismatch / Hetero Region /Consensus Modified info.
-
4
Detailed information on each gene
UTR/Core Exon/Add. Exon/Intron area info.
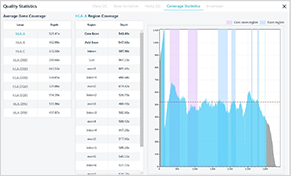
Coverage of the entire genetic area
Hetero position info.
Comparison with other genotypes
-
5
Mapped read check
Mismatch position and info.
Insertion/Deletion position and info.
-
6
General QC result
Review function available
-
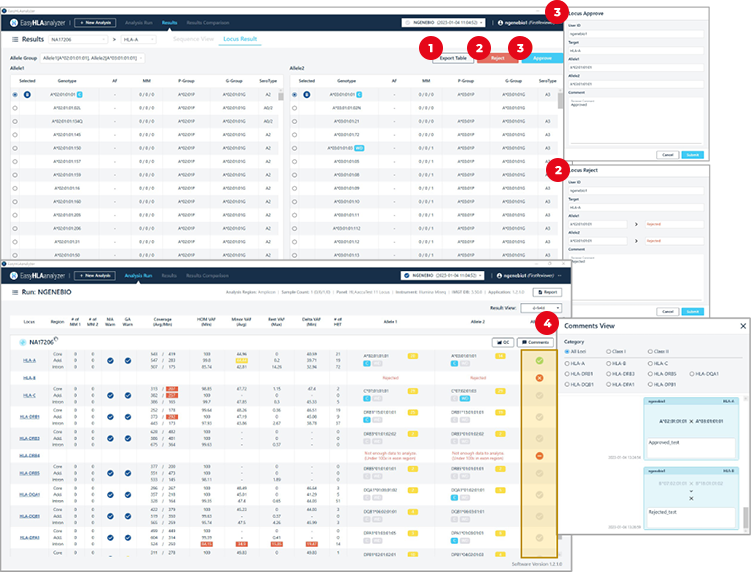
1
Export Table: genotype list (excel)
-
2
Reject: Opinion sharing on rejected results
-
3
Approve: Opinion sharing on approved results
-
4
Checking result approvals/opinions on each gene on the entire sample screen.
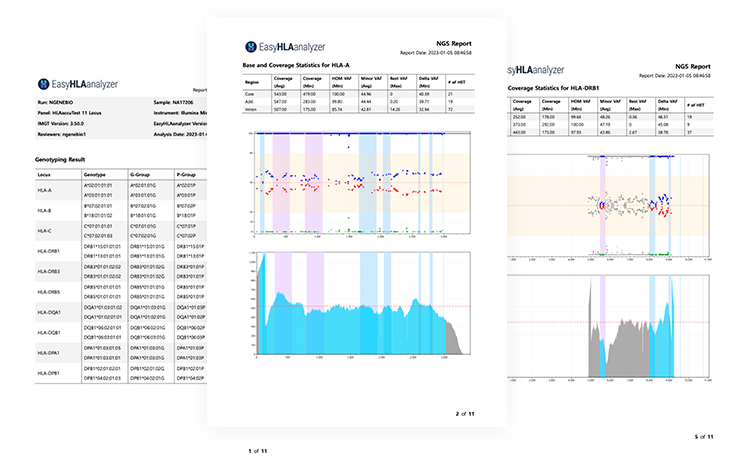
Analysis reports are provided (QC report, clinical report, etc.)
Checking the entire genotype/G-Group/P-Group/Serotype results of analyzed genes
Coverage of each area/VAF info.
Base variation plot/coverage plot info of each gene
WORKING PROCESS
-

- Login
-
SW installation on the user’s PC
Login using ID and PW.
-
- DATA upload
-
Select data from the user’s PC and then upload the analysis
Once uploading is completed, analysis is initiated.
Automatic analysis based on the panel type, IMGT version, and selected analysis area
The current status of the analysis is displayed.
-
- HLA analysis
-
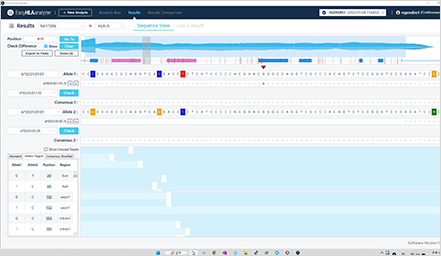
Genotype result check based on the sample and gene
Coverage/VAF check, comparison with other types, and revision if necessary
Based on shared opinions among reviewers, decision-making on the approval of results
-
- Report
-
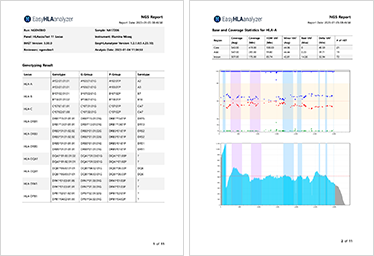
Patient and test information check
Genotype result check for each gene
Coverage/VAF information check for each area
Issuance of report on approved results
Featured compatible panels
EasyHLAanalyzer™ allows you to easily check the individual histocompatibility antigen type analysis results and gene mutation information results.
-

HLAaccuTest™
HLAaccuTest™ All